Structure-guided linker design
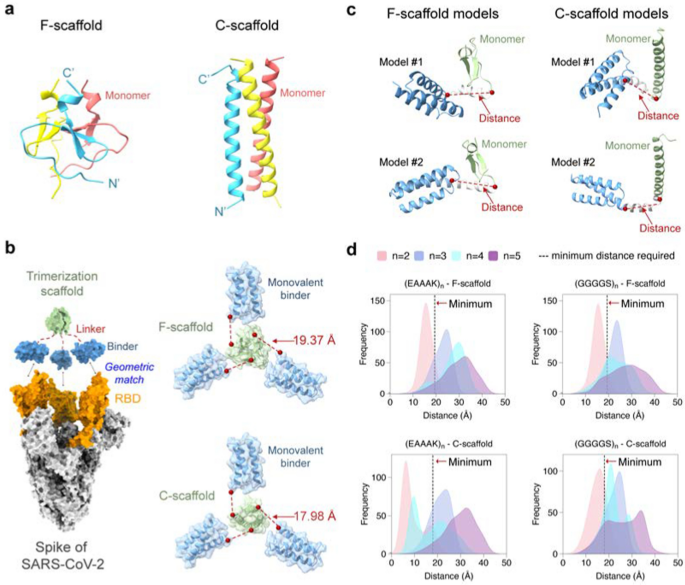
To design a self-assembling trivalent protein with desired physiochemical properties and therapeutic efficacy, we carefully selected two essential components: the monovalent therapeutic agent and the trimerization scaffold. As mentioned above, microproteins are well suited to be engineered into multivalent formats due to their small size, stability, and ease of production. As a proof of concept, we first used the microprotein LCB3 [26] as a monovalent binder to the S protein of SARS-CoV-2. This microprotein (MP) is a mini-mimetic of the ACE2 protein with only 64 aa (hereafter we refer to it as miniACE2) and has been reported to bind to the RBD of the S protein. To obtain optimal multivalent constructs, we then selected two well-studied self-assembling domains as the test trimerization scaffolds, namely the β-propeller-like foldon domain of T4 fibritin used in vaccines [27,28,29] and an α-helical coiled-coil peptide [30], which will be referred to as F-scaffold and C-scaffold, respectively (Fig. 2a).
Structure-guided linker design. a Two trimerization scaffolds used in this study. b The left panel: The trivalent construct designed to match the geometry of the three binding sites on RBDs (orange) of the S protein (silver). The monovalent binder and the trimerization scaffold are shown in blue and green, respectively. The right panel: The calculated minimum distances required between the binder and scaffold. c Schematic of linker models generated by RosettaRemodel. For each model, the distance from the C-terminus of the binder to the N-terminus of the scaffold was determined. d Distributions of the mentioned distances of the designed linkers of different lengths. The dotted black lines indicate the minimum distances required for the constructs
We hypothesized that a well-designed trivalent protein could simultaneously engage all three RBDs of the S protein, thereby blocking the ACE2 binding and enhancing its neutralizing activity against the SARS-CoV-2 variants. It has been shown that RBD can adopt two different conformations: standing-up conformation (RBD-up) for receptor binding and lying-down conformation (RBD-down) for immune evasion [7, 31, 32]. The RBD-up state is essential for membrane fusion and virus entry [9, 31], and potentially facilitates immune clearance. Therefore, our designed goal was to trap the active RBD-up conformation by fully occupying all three RBDs with a designed trivalent protein. To achieve this, we superimposed the miniACE2-RBD complex (PDB ID: 7JZM) onto the S protein with the 3-RBD-up state (PDB ID: 7CT5). Based on the superimposed structure, we calculated the minimum distance required for a linker to connect miniACE2 and the given trimerization scaffold using the Lagrange multiplier method, and found that the minimum distances for the F- and C-scaffolds are 19.37 Å and 17.98 Å, respectively.
Based on the minimum distances, we then designed linkers to connect the monovalent binder and the trimerization scaffold, ensuring an appropriate geometry to match the homo-trimeric target sites. We selected two widely used penta-peptide fragments, the flexible GGGGS and the rigid EAAAK [33], as the building fragment for the candidate linkers, and then determined the repeat number (n) of the given fragment in the linker according to the minimum distances and the folding conformations of each linker. Considering the maximum length of the extended conformation of a penta-peptide (GGGGS or EAAAK), at least two copies of the fragments (i.e., 10-aa length) are needed for a linker to connect the binder to the scaffold. To determine the optimal repeat number n, we used RosettaRemodel to sample a large number of the lowest-energy folding conformations for the linkers of (GGGGS)n or (EAAAK)n (n = 2, 3, 4, or 5) (see Materials and methods). For each n, the folding conformations of the given linker were predicted with the binder at its N-terminus and the scaffold at the C-terminus; and in the calculations, both the binder and the scaffold were treated as rigid bodies. After the conformational sampling, the 1000 top-ranking lowest-energy conformations were used to calculate the distributions of the distances between the C-terminus of the binder and the N-terminus of the scaffold (Fig. 2).
As shown in Fig. 2b, most of the sampling distances for the linkers (GGGGS)2 and (EAAAK)2 fell short of the above-mentioned minimum distances required for the geometric matching of the binder and the trimerization scaffold, indicating that linkers designed with n = 2 may not be suitable. In contrast, most distances for the linkers (GGGGS)n and (EAAAK)n, with n = 3, 4, or 5, exceeded the minimum distances, indicating that n ≥ 3 is required. Among these, the distance distributions for n = 3 were relatively narrow, while those for n = 4 or 5 were wider, indicating that more conformations were energetically possible for these linkers and thus that the binders connected to the trimerization unit could bind to more positions in space, allowing them to adapt their conformations to different epitopes on the target protein. However, the distance distributions for n = 4 were irregular, neither as narrow as n = 3 nor as broad as n = 5; furthermore, those for n = 3 and 5 have covered most of the range seen in n = 4, making n = 4 less preferable. Also, given that n = 5 already covered the possible distances, we no longer explored the situation of n > 5.
Finally, we chose four test linkers, the flexible (GGGGS)3 and (GGGGS)5, and the rigid (EAAAK)3 and (EAAAK)5, to construct the trivalent proteins for miniACE2, resulting in eight trivalent proteins tailored for the two trimerization scaffolds. We designated the two proteins using flexible (GGGGS)n and C-scaffold as MP-3fc, MP-5fc, respectively; those using (GGGGS)n and F-scaffold as MP-3ff, MP-5ff, respectively; those using rigid (EAAAK)n and C-scaffold as MP-3rc, MP-5c, respectively; and those using (EAAAK)n and F-scaffold as MP-3rf, MP-5rf, respectively (Additional file 1: Table S1).
Molecular simulation evaluation of trivalent constructs
Biomedical and therapeutic applications of multivalent proteins usually require them to have good physicochemical properties such as efficient self-assembly and good conformational homogeneity. To identify the best candidate among the eight constructs, we evaluated these properties of the eight constructs using molecular dynamics (MD) simulations. For each construct, three independent simulations were performed, each with a simulation time of 300 ns. Then, we calculated the root mean square deviation (RMSD) of the protein backbone heavy atoms across the simulation trajectories, using their initial structures as the reference conformations (Additional file 1: Fig. S1). As can be seen in (Additional file 1: Fig. S1, the RMSD results showed that all the systems reached equilibrium after about 150-ns simulations. Therefore, we used the post-150 ns trajectories for the following analyses.
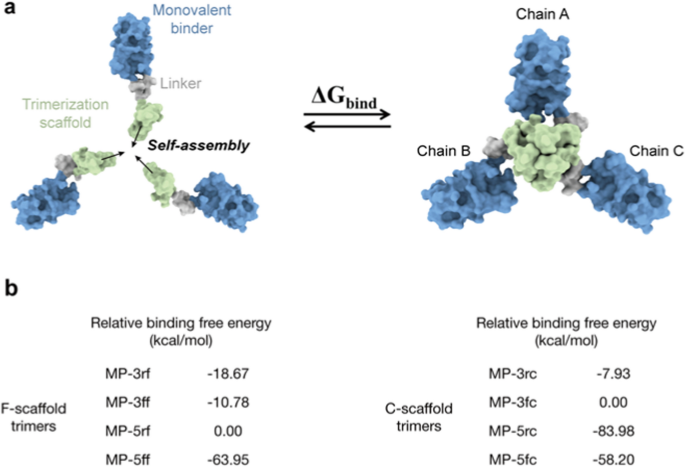
To assess the self-assembly abilities, we first used the Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) method to estimate the binding free energies between the three monomers of the trivalent constructs, as illustrated in Fig. 3a and (Additional file 1: Table. S2). Although MM/GBSA has limitations in predicting absolute values of binding free energy, it excels in ranking the relative binding affinities of different molecules [34]. Similarly, the relative binding free energies of different constructs could also rank their self-assembly abilities. The MM/GBSA calculations showed that the binding free energies of the F-scaffold constructs are typically lower than those of the C-scaffold constructs, except for that of MP-5rc (Additional file 1: Table S2). This suggests that the self-assembly abilities of F-scaffold constructs are relatively better than those of the C-scaffold constructs. Among the F-scaffold constructs, the binding free energy of MP-5rf is the highest; as shown in Fig. 3b, the relative binding free energies of the other three constructs are negative, indicating that their self-assembly abilities are stronger than MP-5rf. Of them, MP-5ff has the lowest relative binding free energy, implying the strongest self-assembly tendency. For the C-scaffold constructs, MP-5rc has the lowest relative binding free energy, indicating a stronger self-assembly ability, especially compared with the higher relative energy values of MP-3fc and MP-3rc.
Molecular simulation assessment of the trimerization tendency of trivalent constructs. a Schematic diagram for the binding process to calculate the binding free energy of a given trivalent construct. b Relative binding free energies of the trivalent constructs calculated by MM/GBSA. For F-scaffold trimers, MP-5rf was used as the reference to calculate the relative values of other constructs. For C-scaffold trimers, the reference is MP-3rc. Please see the MM/GBSA raw data in (Additional file 1: Table S2
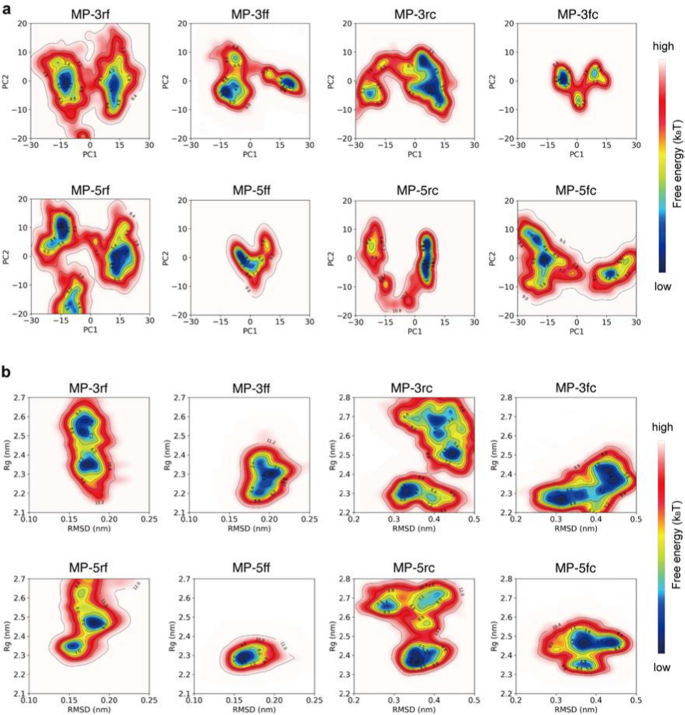
To investigate the conformational homogeneity, we first performed principal component analysis (PCA) on the simulation trajectories and then mapped the simulated conformations of the proteins onto the resulting principal components to generate their free energy landscapes (FELs) (see Methods and materials). As an example, the PCA results for a trajectory of MP-5ff are shown in (Additional file 1: Fig. S2). As seen, the first two principal components contributed to over 80% of the cumulative variance and were thus considered PC1 and PC2 (Additional file 1: Figs. S2A, D). By projecting the simulated conformations onto the two-dimensional space of PC1 and PC2, the resulting FELs showed the distribution patterns of the simulated constructs and their possible numbers of dominant trimer-like conformations in the simulations (Fig. 4).
Free energy landscapes (FELs) for the MD conformations of the trivalent constructs. a FELs of conformational projections onto the first and the second principal components (PC1 and PC2). b FELs of conformational projections onto two alternative reaction coordinates: root mean square deviation (RMSD) and radius of gyration (Rg)
As shown in Fig. 4a, the FEL patterns of the eight constructs are not identical, with about 1–3 low-energy wells (in blue) indicating different numbers of dominant trimer-like conformations in the simulations. Significantly, except for MP-5ff, other constructs had a wider or more than one low-energy well, such as MP-5rc with a wider low-energy well, MP-3rf, MP-5rf with 2 low-energy wells, and MP-3ff, MP-3fc, MP-5fc with 3 low-energy wells. Thus, only MP-5ff showed only a low-energy trimer conformation in the simulations, suggesting that the conformational homogeneity of this protein is the best.
To further confirm the FEL results, we also construct FELs by mapping the simulated conformations onto the two-dimensional space defined by two alternative reaction coordinates: root mean square deviation (RMSD) and radius of gyration (Rg) (Fig. 4b). Consistent with the results of Fig. 4a, except MP-5ff the FELs of all constructs had multiple low-energy wells (in blue), suggesting that multiple low-energy trimer-like conformations coexist in the simulations. Taken together, the results in Fig. 4 suggested that MP-5ff likely has a single stable trimer conformation and thus the best conformational homogeneity.
Experimental validation and functional test
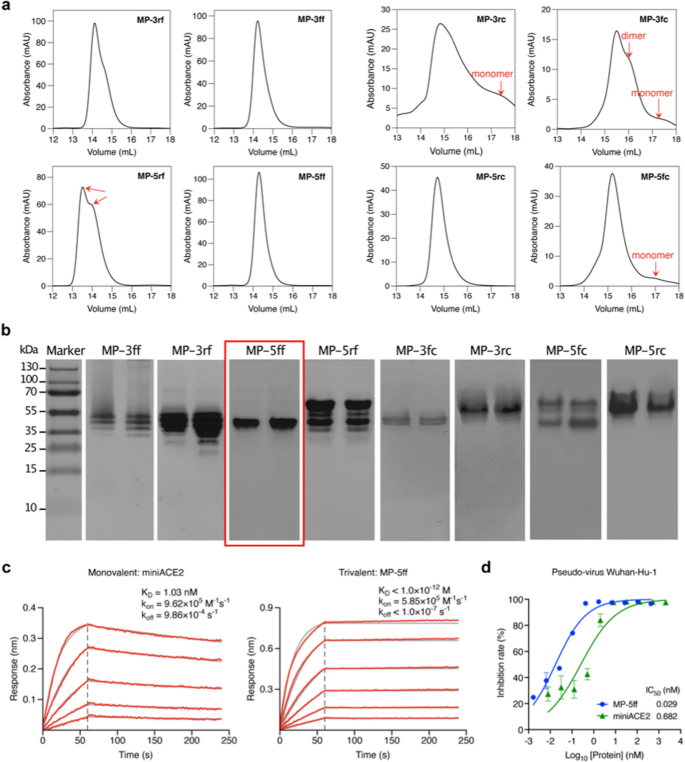
To validate the computational results, we expressed the 8 designed constructs in E. coli Rosetta (DE3) cells and purified the proteins using Ni–NTA affinity chromatography. We then characterized their oligomeric states in solution by size-exclusion chromatography (SEC). As shown in Fig. 5a, the SEC profiles revealed that the four F-scaffold proteins had narrower and sharper trimer peaks than the C-scaffold proteins, indicating a higher trimer ratio in the F-scaffold constructs. Among the F-scaffolds, the peak of MP-5ff appears to be the sharpest and the most concentrated one, indicating that it is the most efficient trivalent construct, in good agreement with the computational evaluation. In contrast, MP-5rf displayed two distinct peaks, probably corresponding to the desired trimerization conformation and another oligomeric state. Indeed, the binding free energy calculations in Fig. 3b have already indicated that the MP-5rf trivalent construct is less stable than the other three F-scaffold constructs. As for the C-scaffold constructs, only MP-5rc had a sharp, single peak indicating a trimer; however, besides the trimer peak, the other three constructs had detectable monomer or dimer peaks, especially MP-3rc and MP-3fc, suggesting that they had a lower trimer ratio. Obviously, these results confirmed the MM/GBSA calculations (Fig. 3b) and showed that those constructs with the lower binding free energies have higher trimerization efficiencies.
Experimental verifications of the trivalent constructs of miniACE2. a SEC profiles of the purified proteins of the constructs. b Native-PAGE analysis of the protein trimer fractions isolated from SEC. c BLI measurements of the binding kinetics of the monovalent miniACE2 and the trivalent construct, MP-5ff, to the immobilized RBD of SARS-CoV-2 (Wuhan-Hu-1). Red traces represent the raw data and the kinetic fits are shown in gray. d Neutralizing activity of miniACE2 and MP-5ff against SARS-CoV-2 pseudovirus (Wuhan-Hu-1)
The FEL analyses in the last subsection have suggested that even in the trimer state, the investigated trivalent constructs are likely to contain several different trimer-like conformations (Fig. 4). To further investigate the possible distributions of trimerization conformations, we performed Native-PAGE analysis on the protein samples collected from the SEC trimer peaks, because this technique can separate two or more trimer-like conformations of the trivalent proteins. As shown in Fig. 5b, among the eight constructs, only MP-5ff presented a single protein band, indicating a single stable trimer conformation, which is consistent with the computational prediction showing only a single energy well in the FELs (Figs. 4a, b). In contrast, MP-3ff and MP-3rf showed multiple distinct bands, indicating that they can adopt several coexisting conformations. For MP-5rf, we observed two dominant bands with several fainter ones at various positions; the upper one may suggest the formation of larger oligomers that fail to maintain a stable trimer. Similarly, MP-5fc also displayed such a pattern. For the other three C-scaffold constructs, we also observed more than one band: MP-3fc exhibited two clear bands, while MP-3rc and MP-5rc showed a clear one and several fainter bands. These findings validate the computational predictions that several low-energy trimer-like conformations may coexist for these constructs (Figs. 4a, b). As a result, MP-5ff was found to be the best construct with the highest trimerization efficiency and conformational homogeneity.
We next examined the target binding affinity of the optimal construct, MP-5ff, to RBD of the S protein using Biolayer interferometry (BLI). Since miniACE2 was originally designed to specifically target the SARS-CoV-2 Wuhan-Hu-1 strain [26], here we focused our functional evaluation on miniACE2 and MP-5ff against this specific strain. As shown in Fig. 5c, MP-5ff exhibited a much slower dissociation rate (koff < 1.0 × 10–7 s−1) compared to that of miniACE2 (koff = 9.86 × 10–4 s−1); thus, the resulting equilibrium dissociation constant (KD) is less than 1 pM, while that of miniACE2 is 1.03 nM. Thus, the binding affinity of MP-5ff for RBD is 1000-fold greater than that of its monovalent counterpart miniACE2, clearly demonstrating that protein multivalency could substantially enhance the target binding affinity. Then, we evaluated the neutralizing activities of miniACE2 and MP-5ff against SARS-CoV-2 pseudovirus (Wuhan-Hu-1). As indicated in Fig. 5d, the monovalent miniACE2 was already able to inhibit the virus with an IC50 of 682 pM; nonetheless, the trivalent MP-5ff still significantly enhanced the neutralizing activity (IC50 = 29 pM), exhibiting a 23-fold increase. Taken together, the trivalent MP-5ff designed by our rational approach has excellent physicochemical properties and potent antiviral activity.
Engineering of a broad-spectrum trivalent nanobody
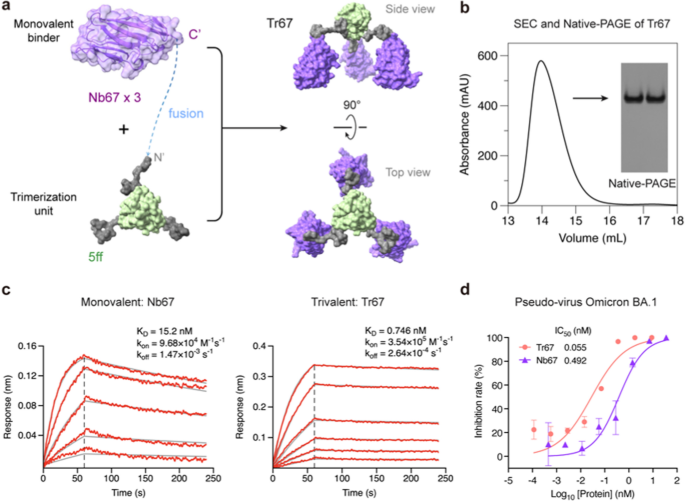
To further demonstrate the effectiveness of our approach, we applied the 5ff trimerization unit to engineer a trivalent nanobody targeting the dominant circulating Omicron variants, because nanobodies represent another widely used category of microproteins well suited for multivalent construction. For this purpose, we selected Nb67, a nanobody identified by Xiang et al. [35] from serially immunized camelid sera, which was reported to neutralize Omicron BA.1. By fusing Nb67 with the 5ff trimerization unit, we created a trivalent nanobody Tr67 (Fig. 6a, (Additional file 1: Table S1). Following the same computational and experimental procedures successfully employed for MP-5ff, we assessed the trimerization efficiency and conformational homogeneity of the engineered Tr67 using MD simulations ((Additional file 1: Fig. S3), SEC and native-PAGE analyses (Fig. 6b). These obtained results demonstrated that Tr67 has a trimerization efficiency and conformational homogeneity very similar to that of MP-5ff.
Design and experimental characterization of Tr67. a Schematic diagram illustrating the trimerization of Nb67 nanobody fused with the optimal trimerization unit 5ff (see its amino-acid sequence of the fusion monomer of Nb67 with 5ff in (Additional file 1: Table S1). b SEC and Native-PAGE analysis of Tr67, showing high trimerization tendency and conformational homogeneity. c BLI measurement of the binding kinetics of the monovalent Nb67 and Tr67 to the immobilized RBD of SARS-CoV-2 pseudovirus (Omicron BA.1). d Neutralizing activities of Nb67 and Tr67 against SARS-CoV-2 pseudovirus (Omicron BA.1). Three independent experiments were performed
We then measured the binding affinity of Tr67 to the target RBD of the S protein using BLI and its neutralizing activity against SARS-CoV-2 pseudoviruses. As shown in Fig. 6c, Tr67 exhibited a higher association rate (kon) and a lower dissociation rate (koff) compared to its monovalent counterpart Nb67. The resulting KD was 0.746 nM, which is an about 20-fold increase in affinity compared to that of the monovalent Nb67 (KD = 15.2 nM). Similarly, Tr67 showed much stronger inhibitory activity against the SARS-CoV-2 Omicron BA.1 pseudovirus than Nb67, with an IC50 of 55 pM versus 492 pM for Nb67 (Fig. 6d). These results demonstrated that the trivalent nanobody has an enhanced potency and thus greater potential to combat viral infections compared to its monovalent counterpart.
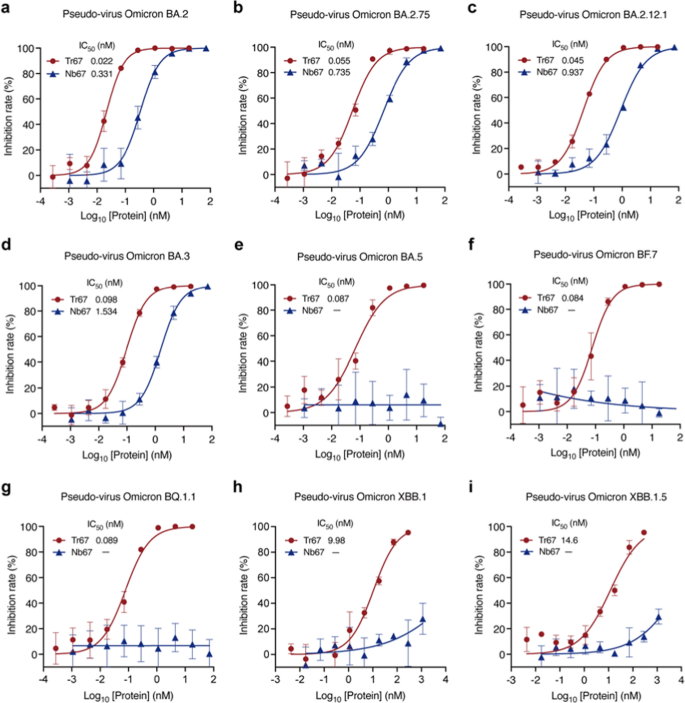
To further investigate the broad-spectrum neutralizing potential of Tr67, we evaluated its neutralization activities against the dominant Omicron variants (Fig. 7). For Omicron BA.2, Tr67 exhibited an IC50 of 0.022 nM and that of Nb67 is 0.331 nM, so the neutralizing activity was greatly enhanced by about 15 folds (Fig. 7a). Similar enhancements were observed for Omicron BA.2.75, BA.2.12.1, and BA.3: the corresponding IC50 values of Tr67 were 0.055, 0.045, and 0.098 nM, respectively, and those of Nb67 were 0.735, 0.937, and 1.534 nM, respectively (Figs. 7b–d). Unexpectedly, Tr67 also neutralized the variants that are more likely to evade humoral immunity. For Omicron BA.5, BF.7, and BQ.1.1, Nb67 failed to achieve any detectable neutralization; however, Tr67 neutralized them with IC50 values of 0.087, 0.084, and 0.089 nM, respectively (Figs.7e–g). Even for the most immune-evasive Omicron XBB family, Tr67 still maintained neutralizing activity, but Nb67 did not (Figs.7h, i). Specifically, the IC50 values of Tr67 against XBB.1 and XBB.1.5 were 9.98 and 14.6 nM, respectively. Thus, compared with its monovalent counterpart, Tr67 has a significant increase in the neutralizing activity against all the tested Omicron variants, suggesting that multivalent proteins have the potential to be developed into broad-spectrum drugs against the emerging SARS-CoV-2 variants.
Broad-spectrum neutralization potential of Tr67 against the dominant SARS-CoV-2 Omicron variants. a—i Neutralizing activities against SARS-CoV-2 pseudoviruses Omicron BA.2, BA.2.75, BA.2.12.1, BA.3, BA.5, BF.7, BQ.1.1, XBB.1, and XBB.1.5 variants, respectively. Three independent experiments were performed for each variant
Cryo-EM analysis of Tr67-spike complex
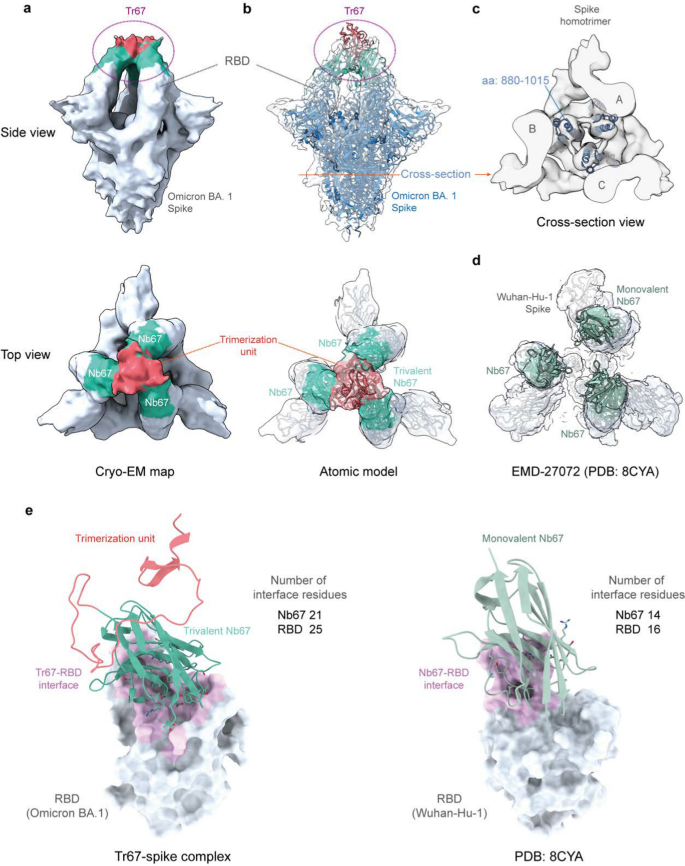
Finally, to confirm whether the binding mode of Tr67 to the spike protein is consistent with our design, we determined the complex structure of Tr67 with the Omicron BA.1 spike protein using cryo-EM ((Additional file 1: Figs. S4, Table S3 and Fig. 8). As seen in Fig. 8A, the cryo-EM density map obtained by the single-particle 3D reconstruction method clearly shows that the complex structure is a triple-symmetric homotrimer; moreover, the density of Tr67 bound to the RBDs at the top of the S-protein is very well defined (Fig. 8a, side view), and the density of the three Nb67 nanobodies and the trimerization unit 5ff (Fig. 8a, top view) can also be distinguished. Therefore, the cryo-EM structure provided experimental evidence that Tr67 is indeed bound to the epitopes specified by the computational design.
Cryo-EM structures of Tr67 in complex with the SARS-CoV-2 (Omicron BA.1) spike protein. a EM density map of Tr67-spike complex in “3-RBD-up” conformation at the overall resolution of 9 Å. b Fitting of the atomic model of the designed Tr67-spike complex into the EM density map. The EM density is shown as a transparent gray surface and the spike protein is rendered in blue. c Cross-section view of the stem region of the spike protein. d An open-like, “3-RBD-up” conformation with the SARS-CoV-2 (Wuhan-Hu-1) spike protein induced by three separated Nb67. e The binding interfaces of trivalent (left panel) and monovalent (right panel) Nb67 binding with the RBDs (shown as gray surface) are shown in pink, and the contact residues on Nb67 are shown as sticks
To obtain the atomic model of the cryo-EM structure, we used the MD flexible fitting method to fit the atomic model of the Tr67-spike complex constructed in the computational design into the density map (Fig. 8b). As can be seen, the atomic model of the whole complex fitted well into the map; and this became clearer by the illustration of the central alpha-helical regions in the S protein stem (Fig. 8c). More specifically, the designed Tr67 matches well with corresponding densities, showing that Tr67 exactly binds to the desired positions on the S protein (Fig. 8b, top view).
Significantly, we found that the complex structure is one in which all 3 RBDs of the S protein are in the standing-up state (3-RBD-up). Due to its amino acid mutations and RBD-RBD interactions, the Omicron spike is usually stabilized in the “2-RBD-down, 1-RBD-up” conformation; this conformational state was considered to facilitate the up-RBD to approach ACE2 and then to promote membrane fusion [36,37,38]. Unlike the spikes of early variants such as Wuhan-Hu-1, the Omicron spike rarely occurs in the 3-RBD-up conformation, which may need to be induced by a combination of distinct antibodies [39, 40]. Thus, the cryo-EM structure did confirm that Tr67 can induce the Omicron spike into the 3-RBD-up conformation. Note that, unlike the common 3-RBD-up conformation (Wuhan-Hu-1) induced by monovalent nanobodies, in which the three RBDs are in an open-like, unassociated state (Fig. 8d), Tr67 has an additional trimerization unit that covalently links the three binders and thereby firmly locks the three RBDs in an inactive state (Fig. 8b, top view). Unsurprisingly, the S protein in such a Tr67-bound state cannot bind ACE2 anymore and therefore its membrane fusion function is completely inhibited.
To understand the molecular basis of the increased binding affinity of Tr67 for the S protein, we analyzed the binding interfaces of monovalent Nb67 and Tr67 with the RBDs using the Nb67-spike structure and the atomic model of the Tr67-spike complex. We identified the interface (contact) residues by a 4-Å distance cutoff between the atoms of Nb67 and those of the RBD, as shown in Fig. 8e and listed in (Additional file 1: Table S4. As can be seen, the binding sites of Nb67 and Tr67 on the RBDs are identical to those of ACE2. However, the Nb67 binder in Tr67 has a larger contact area with the Omicron RBD, and the interface contains 21 residues from Nb67 and 25 residues from the Omicron RBD. In contrast, the monovalent Nb67 and the Wuhan Hu-1 RBD have only 14 and 16 interfacial residues, respectively. In addition, the Nb67 binder in Tr67 forms more hydrogen bonds and salt bridges (Additional file 1: Table S4), suggesting stronger binding interactions. Consistent with the BLI results in Fig. 6c, the number of interfacial residues also supports that Tr67 could establish a more extensive network of interactions, contributing to stronger binding to the Omicron BA.1 and thus enhancing its neutralizing activity.
To explain why Tr67 also binds to other Omicron variants (Additional file 1: Fig. S5), we built structural models for their RBDs based on the atomic models in Fig. 8b, and then analyzed their binding interfaces with the monovalent Nb67 and Tr67 by docking simulation using PyDock [41]. The best-scoring binding poses from the simulations were used as the representatives of the Nb67-spike and Tr67-spike complexes, as shown in (Additional file 1: Figs. S6 and S7, respectively. As illustrated in (Additional file 1: Fig. S6a, for the variants BA.1, BA.2, BA.2.75, BA.2.12.1, and BA.3 (cluster 1), Nb67 was successfully docked into the expected epitope on RBD; however, for variants BA.5, BF.7, BQ.1.1, XBB.1, and XBB.1.5 variants (cluster 2), Nb67 in the best-scoring poses was not located at the expected sites, but at other sites that are sterically unfavorable in the 1-RBD-up conformation of the S protein (Additional file 1: Fig. S6b). Consistent with this, Nb67 was able to neutralize the Omicron variants in cluster 1 but not those in cluster 2 (Fig. 7). Obviously, the amino-acid mutations of the cluster 2 variants weaken the interactions of the monovalent Nb67 with the variant RBD sites in Fig. 8e and thus abolish the neutralization. Particularly, similar to a previous study [35], we found that the mutation at 486 plays a key role in this process (Additional file 1: Fig. S5 and Table S5). In contrast, for all variants, Tr67 of the best-scoring poses binds to the same epitopes as that of BA.1. The binding interfaces are also larger than those of Nb67 (Additional file 1: Table S6). It appears that the synergistic binding of the three Nb67s in Tr67 increases the binding interface and then leads to higher binding affinities that could resist the mutations such as that at 486 to some extent. As a result, Tr67 is still able to bind to the same epitopes of the Omicron BA.1 S protein and neutralize the other variants tested. Clearly, further structural studies are needed to elucidate the detailed molecular mechanisms involved.